Ashwin Kaliyaperumal
Quantum Combinatorial Optimization Intern | Seattle, Washington, USA
About Ashwin
Hi, I’m Ashwin! I’ve been super fascinated with quantum computing for the past five years, especially with practical quantum algorithms for current, real-world applications. I’ve been immersed with quantum computing ever since Qiskit’s Qubit x Qubit program for high schoolers in 9th grade. My favorite exploration of quantum computing came when I conducted research on a quantum optimization algorithm for efficiently developing bus routes for urban transit planning, which I tested on downtown Seattle’s bus stop grid.
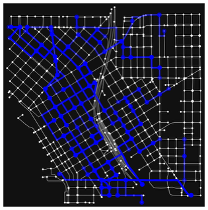
At UW, I co-founded UW’s Quantum Computing Club and have hosted two Qiskit Fall Fests, where we taught undergraduates about quantum computing - the theory, software, and hardware behind it. I’ve also presented my quantum urban transit research at QED-C’s Plenary in 2024.

At my internship with IFF Technologies, I’ve been working on a literature review of combinatorial optimization for pharmaceutical companies. I’m focusing on four main use cases where combinatorial optimization formulations (and potential quantum solutions to these formulations) could provide speedups over mainstream solutions: molecular docking, drug synergy/antagonism prediction, pharma supply chain optimization, and hospital supply storage optimization. For each use case, I’m reviewing current classical solutions to these problems, approaches big pharmaceutical companies are taking with AI and LLM’s, and existing literature for any potential quantum computing solutions (both gate-based and annealing) to these problems.
I’ve also been working on a research paper comparing quantum annealing approaches to solve the Molecular Docking problem with graph-subgraph isomorphism. I’m creating a new QUBO formulation for the Molecular Docking problem, based on the work of Triuzzi et. al., and I’ve been comparing the performance of my formulation, the formulation from Triuzzi et. al., and a graph-subgraph specific QUBO formulation from Calude, Dinneen, and Hua's "QUBO formulations for the graph isomorphism problem and related problems". I’m also categorizing the scale of problems for the Molecular Docking problem through quantifying the distribution of PDBBind+ protein-ligand complexes to better understand the technical limitations of both quantum algorithms run on both current and future quantum hardware.

Please get in touch with me if you’re interested in my work! My research is to be published soon, so stay in the loop by adding me on Linkedin !